RADIANT (beta)
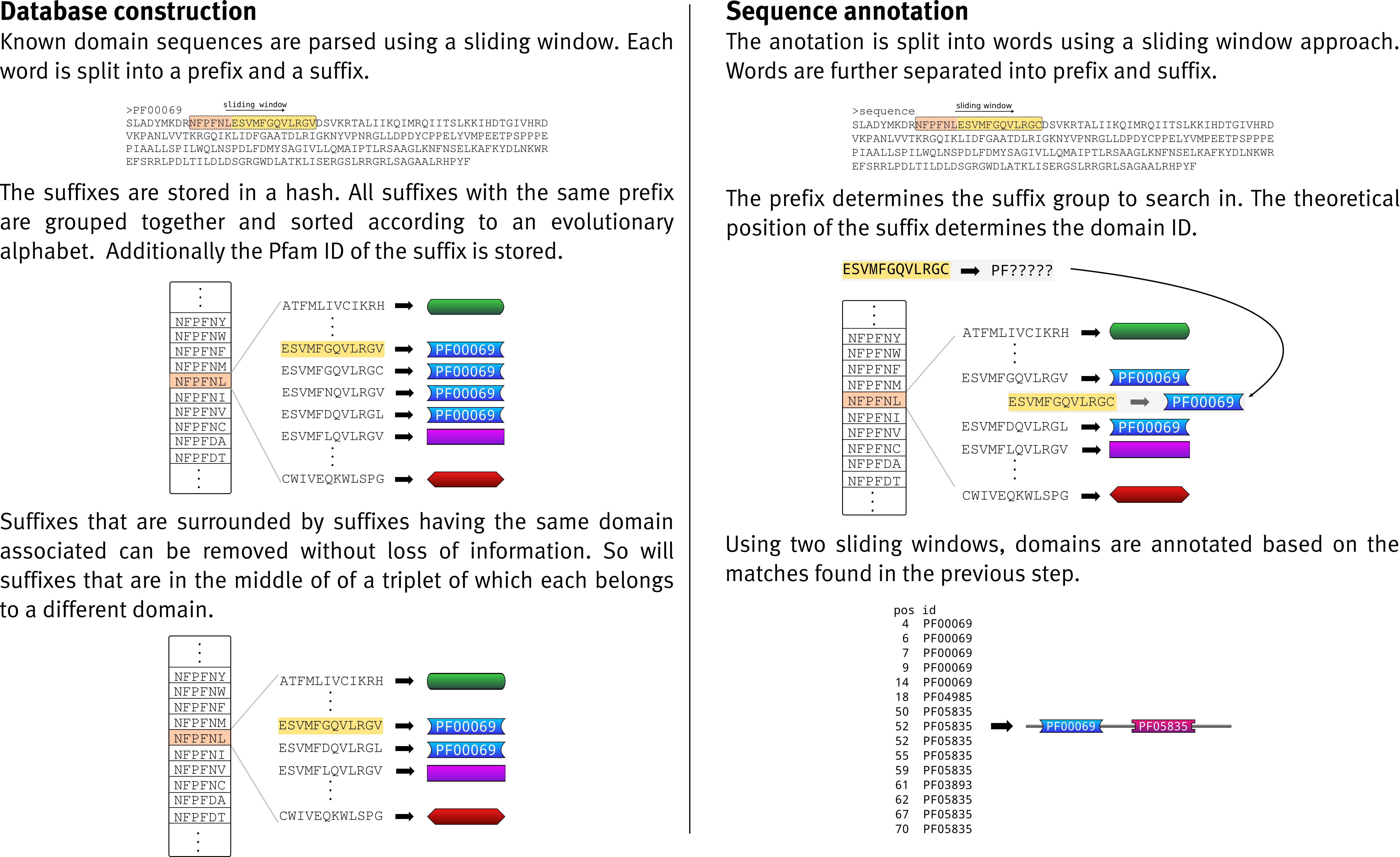
RApid DomaIn ANnoTation is a program for fast domain annotation of protein as well as assembled transcriptome data. It is based on the ideas implemented in UProC but has been expanded so that it is able to determine the order of domains in the arrangement. Although, it is still under development the results we have are very promising.
Download
There are two ways to download RADIANT. Either you can download it using git or manually from the website. Both ways are described below.
Tip
If you use git you can easily update to a newer version when available.
Download using git
Use git to clone the repository and download BioSeqDataLib as a submodule:
git clone https://zivgitlab.uni-muenster.de/domain-world/RADIANT.git
cd RADIANT
git submodule init
git submodule updateManual Download
You can download the compressed source code from our gitlab web-page https://zivgitlab.uni-muenster.de/domain-world/RADIANT. Additionally you will have to download the BioSeqDataLib from https://zivgitlab.uni-muenster.de/domain-world/BioSeqDataLib and uncompress it in the libs directory of RADIANT.
Compilation & Installation
Inside the source folder a build directory is needed in which the code will be compiled. CMake is used to find all the needed requirements of the library.
mkdir build
cd build
cmake ..
makeUpdating
Sometimes it will be necessary to update RADIANT either because it contains some new features or because we unfortunately had a bug somewhere that we have now fixed. If you used git for the original download you can simply use git to update your code. Simply change into the RADIANT directory and type:
git pull
git submodule foreach git pull origin masterYou now simply follow again the steps in the Installation section. If you downloaded the code without git you will have to download the latest version and replace the old one with it. Do not forget to update the BioSeqDataLib folder as well.
RADIANT data bases
These are the databases compatible with the current version of RADIANT:
| database | version | file | unzipped size |
|---|---|---|---|
| Pfam | 36 | radiant_db_pfam36.tar.bz2 | GB |
| Pfam | 35 | radiant_db_pfam35.tar.bz2 | 7.8 GB |
For older Pfam-Versions and previous versions of RADIANT see here for additional databases: here.
Usage
RADIANT needs two input arguments. One is the proteome or transcriptome to annotate, the second is the database to use. For the database only the prefix needs to be given.
radiant -i proteome.fa -d <path to database>/pfam36 -o annotation.txtOutput
The output of RADIANT is designed to be similar to the PfamScan output but contains less columns. Lines starting with a
‘#’ are comments and contain information about the program and format version and as well about the query and database files
used.
# RADIANT 1.0.0
# RADIANT output format: 1.0
# run at Wed Aug 2 16:24:46 2017
#
# Options used:
#-----------------------------------------------------------------------------
# query file: /global/scratch/ckeme_01/radiant_data/human/Homo_sapiens.GRCh38.pep.all.fa.iso.fa
# database file: /global/scratch/ckeme_01/radiant_data/dennis/pfam31
# translate: false
#-----------------------------------------------------------------------------
#
# <seq id> <match start> <match end> <hmm acc> <hmm name> <type> <clan>
ENSP00000362111.4 27 27 PF00335 Tetraspannin Family CL0347
ENSP00000362122.4 120 120 PF04089 BRICHOS Domain No_clan
ENSP00000360640.4 36 36 PF00535 Glycos_transf_2 Family CL0110
ENSP00000356744.1 42 42 PF00069 Pkinase Domain CL0016
ENSP00000352276.4 184 184 PF14868 DUF4487 Family No_clan
ENSP00000363117.3 91 91 PF00018 SH3_1 Domain CL0010
ENSP00000363117.3 152 152 PF00017 SH2 Domain CL0541
ENSP00000363117.3 272 272 PF07714 Pkinase_Tyr Domain CL0016Restrictions
The database is based on the Pfam-A.fasta file from the Pfam website. Unfortunately not all domains are contained in this file. Therefore some domains will never be annotated. Furthermore, very short domains and domains that are very similar to others will most likely not be annotated either.
Contact the developer
If you find a problem, have questions or any kind of comment please contact us (domainworld[@]uni-muenster.de).