DomRates
Modularity is an important keystone in molecular evolution and indispensible for evolutionary innovation. Protein domains as the modules of proteins can be reused in different molecular contexts and therefore rearrangements of domains can create a broad functional diversity with just a few mutational steps. DomRates infers these rearrangement events of protein domains for a given phylogeny and calculates the rates of the related events.
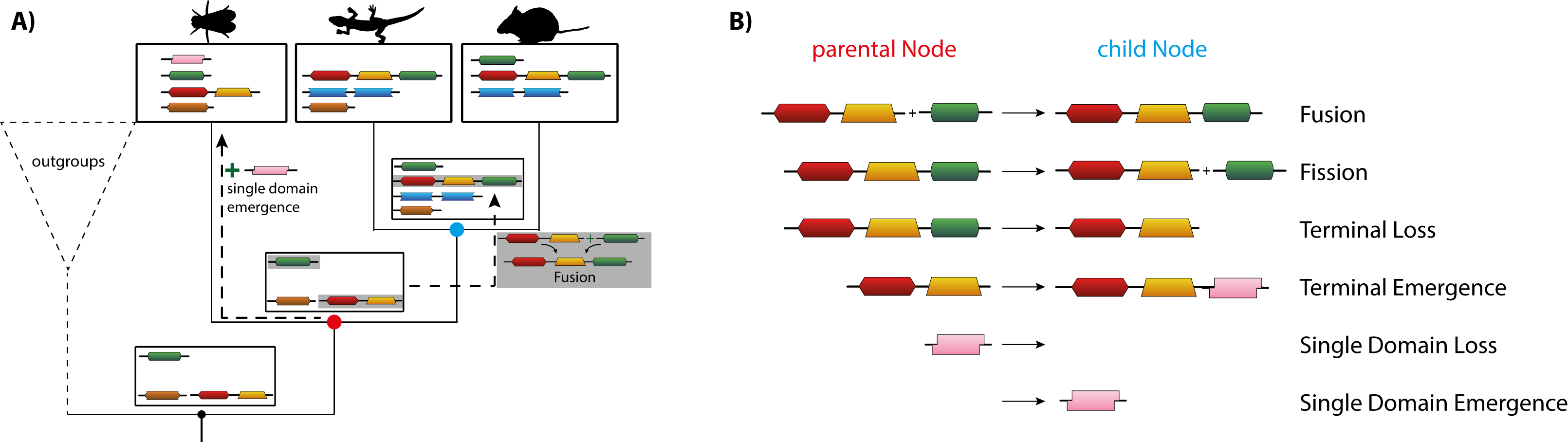
DomRates Illustration: (A) shows the Inference of different rearrangement events for an example phylogeny with different insect species via DomRates. (B) lists all six different single-step events DomRates takes into account.
This website covers the basics of DomRates. For more detailed information please check the UserManual.
Download & Setting up
Requirements
Although we try to keep the dependencies to a minimum some existing libraries are needed.
- cmake (https://cmake.org)
- compiler supporting c++11 - e.g. g++ 4.8 or higher
- boost modules: filesystem, program_options (http://www.boost.org/)
- BioSeqDataLib (see download instructions)
Optional:
- git (https://git-scm.com) Recommended: It simplifies future update processes
In most Linux distributions (e.g. Ubuntu, Arch Linux) it should be possible to install most of these dependencies using the package manager.
Download
There are two ways to download DomRates. Either you can download it using git or manually from the website. Both ways are described below.
Tip
If you use git you can easily update to a newer version when available.
Download using git
Use git to clone the repository and download BioSeqDataLibrary as a submodule:
git clone https://zivgitlab.uni-muenster.de/domain-world/DomRates.git
cd DomRates
git submodule init
git submodule updateManual Download
You can download the compressed source code from our gitlab web-page https://zivgitlab.uni-muenster.de/domain-world/DomRates. Additionally you will have to downolad the BioSeqDataLib from https://zivgitlab.uni-muenster.de/domain-world/BioSeqDataLib and uncompress it in the libs directory of DomRates.
Compilation & Installation
Inside the source folder a build directory is needed in which the code will be compiled. CMake is used to find all the needed requirements of the library.
mkdir build
cd build
cmake ..
makeUpdating
Sometimes it will be necessary to update DomRates if it contains some new features or because we unfortunately had a bug somewhere that we have now fixed. If you used git for the original download you can simply use git to update your code. Simply change into the DomRates directory and type:
git pull
git submodule foreach git pull origin masterYou now simply follow again the steps in the Installation section. If you downloaded the code without git you will have to download the latest version and replace the old one with it.
Running DomRates
After you have set up your system as described above you can get a short overview on how to use DomRates by displaying the help.
./domRates --helpA more detailed description can be found in the manual.
A standard way of analysing your data with DomRates would be:
./domRates -t tree.nwk -a /dir/with/annotation-files/ -g outgroup -o rates.txt -s statistics.txtThe provided tree (tree.nwk) has to be a bifurcating tree in newick format and the names of the leaves fit to the names of the files in the directory with annotation files (-a). The name of the outgroup in the tree can be provided with the -g option and -o specifies the name for the general output file with all rates, while -s specifies a name for text files with additional statistics.
Visualization of Results
To visualize some of the results produced by DomRates there is a python script in the src folder of DomRates to process the statistics result file.
The different parameters are explained in the help.
python visualization_domrates_tree.py --helpA standard way of analysing your results produced with DomRates as described above can be achieved via:
python visualization_domrates_tree.py -t tree.nwk -s statistics.txt -o outputPDFThis would result in a outputPDF file showing a visualization of the distribution of all events over the given phylogeny like in the example figure below.
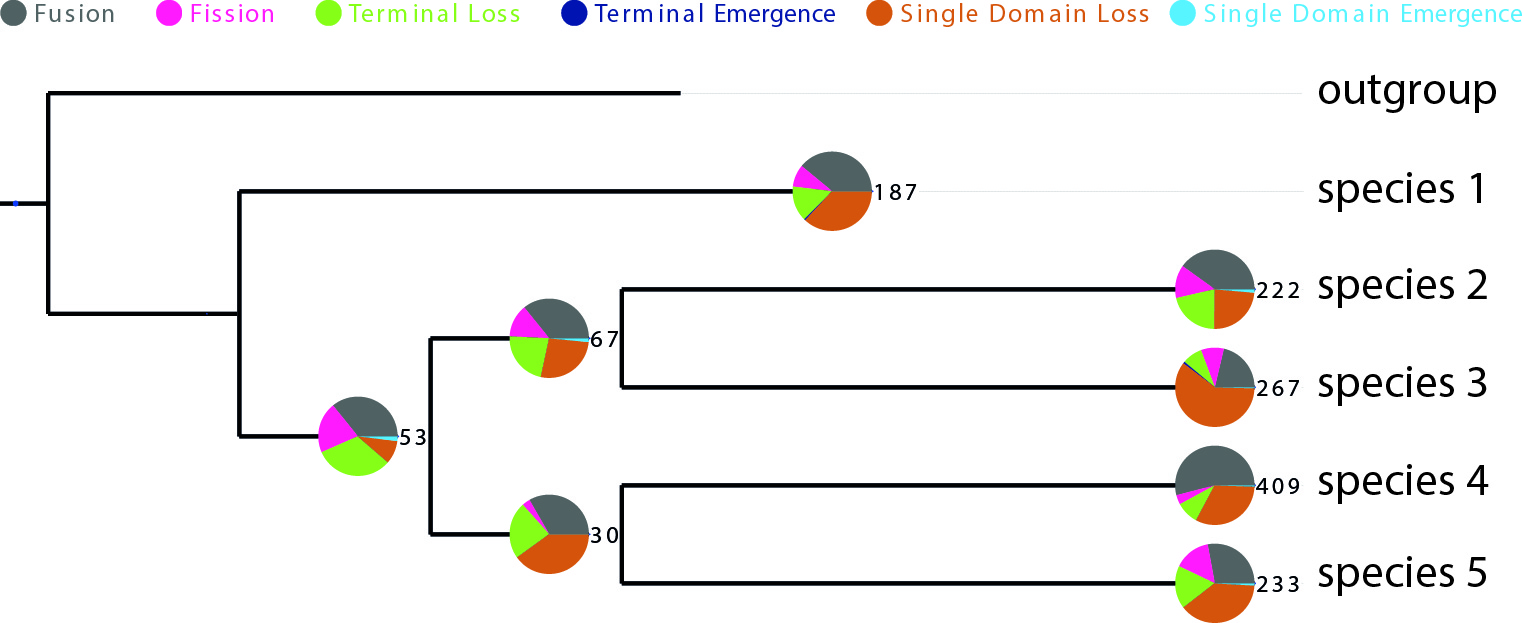
DomRates Results Visualization
To get information about all different options please check the manual.
Contact the developer
If you find a problem, have questions or any kind of comment please contact us (domainworld[@]uni-muenster.de).
Citation
If you use DomRates in your project please cite our publication:
Dohmen E, Klasberg S, Bornberg-Bauer E, Perrey S, Kemena C, The modular nature of protein evolution: domain rearrangement rates across eukaryotic life, BMC Evolutionary Biology (2020), doi: 10.1186/s12862-020-1591-0
https://bmcevolbiol.biomedcentral.com/articles/10.1186/s12862-020-1591-0
Example publications utilizing DomRates
- Thomas GWC, Dohmen E, Hughes DST, et al., Gene content evolution in the arthropods, Genome Biology, 2020. https://doi.org/10.1186/s13059-019-1925-7